Structural cells are key regulators of organ-specific immune responses.
Thomas Krausgruber* & Nikolaus Fortelny* et al.
Nature (2020), doi: 10.1038/s41586-020-2424-4
For further context, please see the following News & Views articles:
An antiviral response beyond immune cells.
Tomás Gomes & Sarah A. Teichmann
Nature (2020), doi: 10.1038/d41586-020-01916-2
A gene atlas of ‘structural immunity’.
Kirsty Minton
Nature Reviews Immunology (2020), doi: 10.1038/s41577-020-0398-y
Organ immune responses — don’t forget the structural cells.
Zewen Kelvin Tuong & Menna R. Clatworthy
Nature Reviews Nephrology (2020), doi: 10.1038/s41581-020-00334-x
Study overview
Multi-omics profiling establishes cell-type-specific and organ-specific characteristics of structural cells.
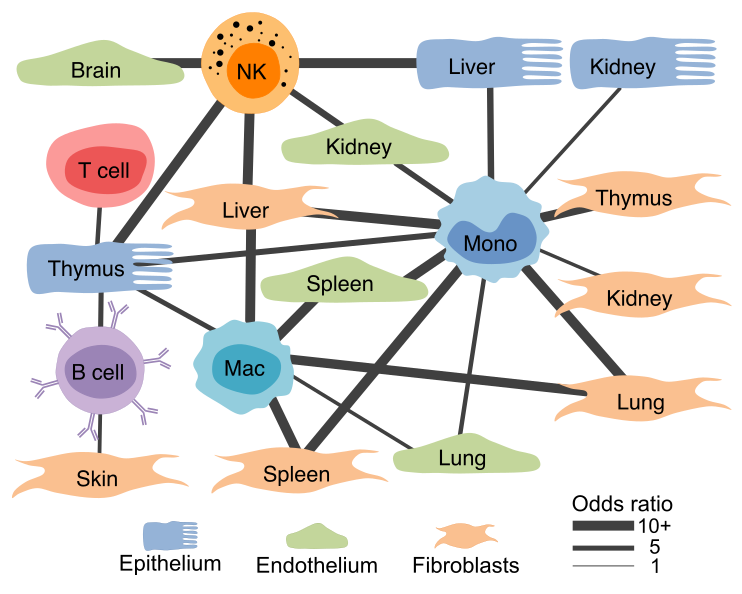
Gene expression profiles of structural cells predict cell-type-specific crosstalk.
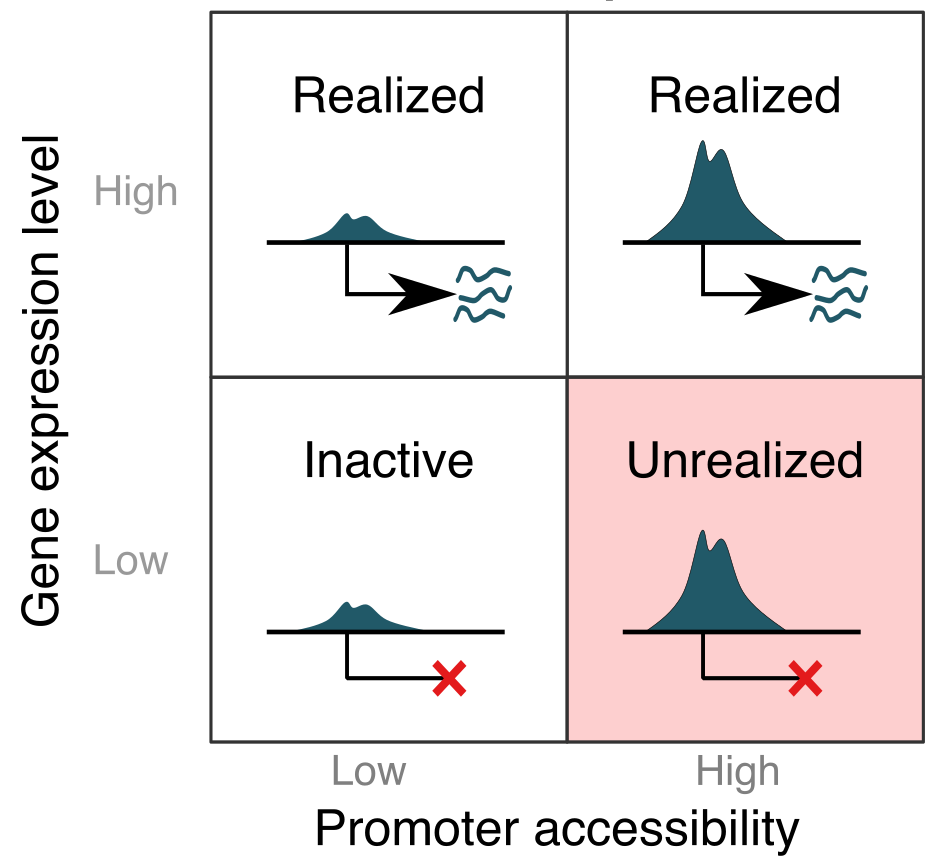
The immunological potential of structural cells is epigenetically encoded.
Response to viral infection realizes the immunological potential of structural cells.
Data
This section provides access to raw data and analysis results.
| Description | Links |
|---|---|
|
Genome (mm10) browser tracks: |
All samples and assays, fully customizable: UCSC Track Hub Aggregated ATAC-seq tracks: Link Aggregated ChIPmentation tracks: Link Aggregated RNA-seq tracks: Link Aggregated RNA-seq tracks of cytokine treatments: Link |
|
Sequencing data (Uploaded to Gene expression omnibus - GEO) |
Superseries across all data (GEO SuperSeries GSE134663) ATAC-seq data (GEO Series GSE134648) ChIPmentation data (GEO Series GSE134658) RNA-seq data (GEO Series GSE134659) RNA-seq data of cytokine treatments (GEO Series GSE145820) |
| Results: |
(Supplementary Table 1)
Sequencing statistics of this dataset
(XLS)
(Supplementary Table 2) Expressed marker genes (XLS) (Supplementary Table 3) Cell-cell interactions network at homeostasis (XLS) (Supplementary Table 4) Categorized list of immune-related receptors and ligands (XLS) (Supplementary Table 5) Counts of structural cells in the Tabula Muris dataset (XLS) (Supplementary Table 6) Epigenomic marker regions (XLS) (Supplementary Table 7) Genes with epigenetic potential (XLS) (Supplementary Table 9) Differential genes upon LCMV infection (XLS) (Supplementary Table 8) Curated list of immune-related gene sets used for enrichment analysis (XLS) (Supplementary Table 10) Changes in cell-cell interactions network upon LCMV infection (XLS) (Supplementary Table 11) Differential genes upon Cytokine treatments (XLS) |
| Code: |
All code relevant for data processing and analysis (ZIP archive) |
Citation
If you use these data in your research, please cite:
Thomas Krausgruber*, Nikolaus Fortelny*, Victoria Fife-Gernedl, Martin Senekowitsch, Linda C. Schuster, Alexander Lercher, Amelie Nemc, Christian Schmidl, André F. Rendeiro, Andreas Bergthaler and Christoph Bock.
Structural cells are key regulators of organ-specific immune responses.
Nature (2020), doi: 10.1038/s41586-020-2424-4
* These authors contributed equally to this work
Correspondence: cbock@cemm.oeaw.ac.at (C.B.)